Characterizing neuronal transcriptome and RNP complex changes in SMA and ALS models
The known roles of SMN, TDP-43 and FUS in RNA metabolism suggest that mutations in these proteins may affect the expression of specific genes and mRNA isoforms. However, a detailed description of transcriptome changes in relevant disease models is still lacking.
In this project, we aim to identify and validate common gene expression and ribostasis pathways underlying motor neuron degeneration in ALS SMA in vivo in adult onset fly disease models and to answer these specific questions:
- Which genes and splicing isoforms display altered expression in the neuronal tissue of ALS and SMA Drosophila models?
- What are the neuronal-specific RNA-protein and protein-protein interactions established by the Drosophila homologues of disease causing proteins Fus, TDP-43 and SMN?
- Are there common neuronal gene networks or pathways in these disease models and what is their contribution disease progression?
- Are the candidates identified in Drosophila misregulated in human disease patients?
To achieve these aims, we are developing late-onset fly ALS and SMA models targeting the TDP-43, FUS and SMN homologues and generating high quality RNA-seq neuronal transcriptome and RNA-protein interaction profiles. The information contained in this dataset will be explored using computational approaches that integrate additional data available within the consortium and in public repositories to identify altered gene interaction networks and pathways, which will be mapped to the human counterpart. Finally, results will be functionally validated in flies and human patient samples to identify clinically relevant markers and therapeutic targets.
Results
Connecting SMA and ALS through protein interactions
We performed a systematic literature review that collects all the most recent understanding of key biomolecular processes that underlie MNDs focusing on the FUS, TDP43, SMN and SETX proteins. This review was complemented with a systems approach to explore the interactomic and functional framework of MN degeneration. Using several publicly available protein-protein interaction (PPI) databases and literature we constructed a focused PPI network around these three proteins to characterize their molecular environment and thus, identify their most critical functions within this context. This analysis revealed a highly interconnected network of interactions between the four proteins, foreshadowing the most recent discoveries linking them to transcription termination and DNA repair.
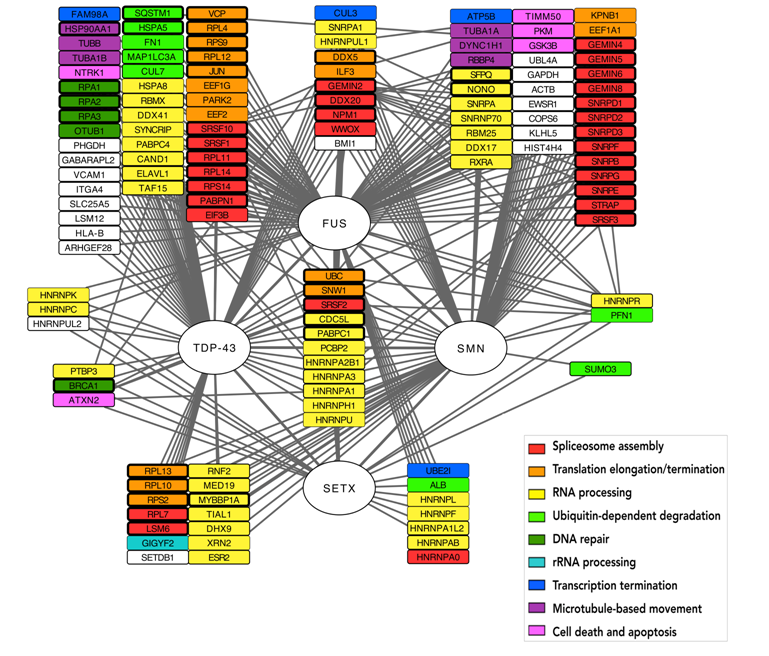
A computational method to identify genes connecting SMA and ALS
We have developed a new computational method based on a new measure of neighborhood within PPI networks for identification of genes that act as connecting hubs between two diseases. We expect this method will identify novel MN disease-related gene candidates to be experimentally tested in fly models and human patient samples. This method may be expanded to other diseases that share common molecular pathways.
References
- Miguel-Aliaga I, Chan YB, Davies KE, Van Den Heuvel M. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett. 2000;486(2):99–102.
- Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121(10):4118–26.
- Kitano H. Systems biology: A brief overview. Science (80- ). 2002;295(5560):1662–4.
- Zaldívar T, Montejo Y, Acevedo AM, Guerra R, Vargas J, Garofalo N, et al. Evidence of reduced frequency of spinal muscular atrophy type I in the Cuban population. Neurology. 2005;65(4):636–8.
- Simic G. Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 2008;116(3):223–34.
- Siddique N, Siddique T. Genetics of Amyotrophic Lateral Sclerosis. Phys Med Rehabil Clin N Am. 2008;19(3):429–39.
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–65.
- Carrì MT, Valle C, Bozzo F, Cozzolino M. Oxidative stress and mitochondrial damage: importance in non-SOD1 ALS. Front Cell Neurosci. 2015;9:41.
- Cooper TA, Wan L, Dreyfuss G. RNA and Disease. Cell. 2009;136(4):777–93.
- Sun S, Ling S-C, Qiu J, Albuquerque CP, Zhou Y, Tokunaga S, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun. 2015;6:6171.
- Tsuiji H, Iguchi Y, Furuya A, Kataoka A, Hatsuta H, Atsuta N, et al. Spliceosome integrity is defective in the motor neuron diseases ALS and SMA. EMBO Mol Med. 2013;5(2):221–34.
- Yamazaki T, Chen S, Yu Y, Yan B, Haertlein TC, Carrasco MA, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2(4):799–806.
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human Senataxin Resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-Dependent termination. Mol Cell. Elsevier Inc.; 2011;42(6):794–805.
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. Nature Publishing Group; 2016;529(7584):48–53.
- Suraweera A, Lim YC, Woods R, Birrell GW, Nasim T, Becherel OJ, et al. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet. 2009;18(18):3384–96.
- Bennett CL, La Spada AR. Unwinding the role of Senataxin in neurodegeneration. Discov Med. 2015;19(103):127–36.
- Barabási A, Gulbahce N, Loscalzo J. Network Medicine: A Network-based approach to human disease. Nat Rev Genet. 2011;12(1):56–68.
- Kann MG. Protein interactions and disease: Computational approaches to uncover the etiology of diseases. Brief Bioinform. 2007;8(5):333–46.
- Calderone A, Castagnoli L, Cesareni G. mentha: a resource for browsing integrated protein-interaction networks. Nat Methods. Nature Publishing Group; 2013;10(8):690–1.
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:214–20.
- Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, et al. The MIntAct project - IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42(D1):358–63.
- Aibar S, Fontanillo C, Droste C, De Las Rivas J. Functional gene networks: R/Bioc package to generate and analyse gene networks derived from functional enrichment and clustering. Bioinformatics. 2015;31(10):1686–8.